scikit learn is a great machine learning library for Python. It offers broad range of machine learning algorithms and tools. Learning curves are a new tool that has been merged last week and I want to use that feature to point out why scikit learn is such a great library.
A learning curve shows the training scores and the test scores for varying number of training samples. It is a tool that can be used to find out whether we benefit from more training samples. At a first look, it seems to be very easy to implement. We could do something like:
estimator = ...
X_train, y_train, X_test, y_test = split(X, y)
n_samples = X_train.shape[0]
train_scores, test_scores = [], []
for n in range(10, 10, n_samples):
estimator.fit(X_train[:n], y_train[n])
train_scores.append(estimator.score(X_train[:n], y_train[n]))
test_scores.append(estimator.score(X_test, y_test))
plot(range(10, 10, n_samples), train_scores)
plot(range(10, 10, n_samples), test_scores)
However, there are some flaws in the code that make the implementation actually much worse than the version that is implemented in scikit learn:
- We have to define the split function.
- Actually, real cross-validation would be even better to generate more
stable learning curves. But that would require an additional loop and
afterwards averaging over the runs. (Fortunately, scikit learn could
generate the cross-validation splits for us:
sklearn.cross_validation.) - If we would like to use another scoring, we would have to change that
in the code. Fortunately, scikit-learn comes already with a wide range
of predefined scorers in
sklearn.metrics. - We can parallelize over cross-validation runs and different numbers of training samples and that would require even more code.
- Incremental learning could be exploited! If we would like to train an
online learning algorithm like
sklearn.linear_model.SGDClassifieron a large dataset, we can reuse models that have been fitted on parts of the training set to speed up the generation of the learning curve. - This works only for supervised learning. We can of course compute scores for unsupervised learners like clustering algorithms but that would require additional code.
- Maybe we are not interested in the exact number of training examples but want something like a plot for 10%, 20%, ..., 100% of the training examples.
The three required arrays that are used to actually plot the curves can be
generated in just one call to sklearn.learning_curve.learning_curve
without all these flaws.
Lets take a look at the signature of the function learning_curve:
train_sizes_abs, train_scores, test_scores = \
learning_curve(estimator, X, y,
train_sizes=np.linspace(0.1, 1.0, 10),
cv=None, scoring=None,
exploit_incremental_learning=False,
n_jobs=1, pre_dispatch="all", verbose=0)
It takes an estimator and the whole training set (X, y) where y
could be None. We already have a good default for train_sizes:
an array with dtype == float is interpreted as fractions of the
maximum number of training examples, i.e. the default is like
10%, 20%, ..., 100%. Otherwise the sizes are interpreted as absolute
numbers of training samples. We can set some standard parallelization
options for joblib like n_jobs and pre_dispatch and we can
even exploit incremental learning.
But the magic happens in the arguments cv and scoring. These
parameters rely on the huge feature range of scikit learn. cv can be
a cross-validation generator like sklearn.cross_validation.KFold,
a list of train and test indices for each test run or just an integer
that is interpreted as the number of folds for k-fold cross-validation.
If the estimator is a classifier it will be interpreted as the number
of folds for stratified k-fold cross-validation. scikit learn supports
even more cross-validation methods like leave-one-out CV etc. The default
value (None) will be interpreted as 3-fold cross-validation.
Similar things happen with the scoring parameter. If it is None, the
function assumes that there is a member function estimator.score. We can
also pass a callable with the signature scorer(estimator, X[, y]) or a
string that will be mapped to a built-in scorer like for example r2,
accuracy or precision_scorer. Note that the scorer could also compute
scores for unsupervised learners.
With only a few lines of code we can now plot beautiful learning curves like these:
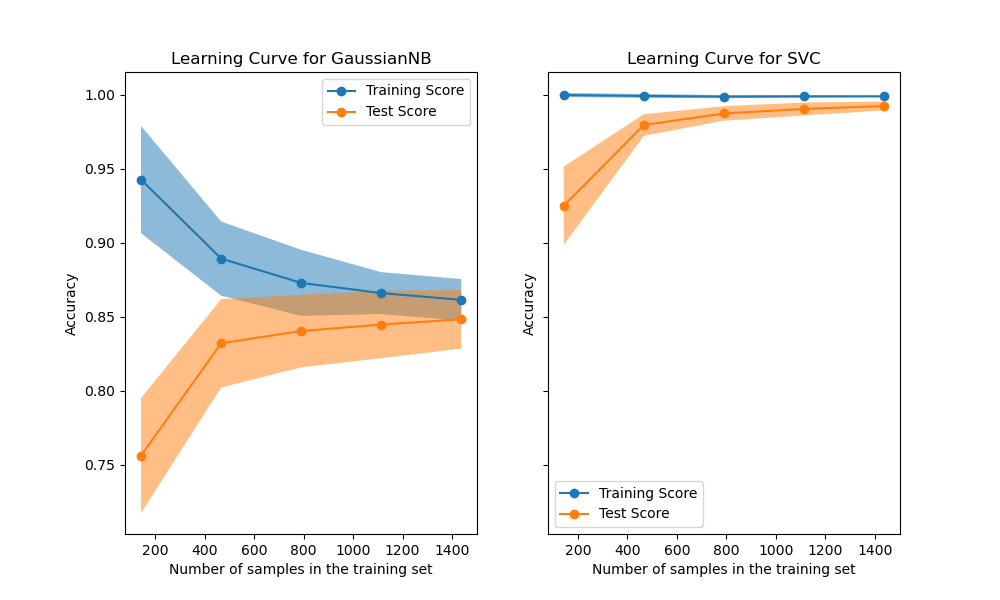
scikit learn offers many more tools like this, for example model selection with grid search or random search, a pipeline that combines preprocessing and learners, simple dataset access and dataset generators and a really great explanatory documentation.